ACTINONIN
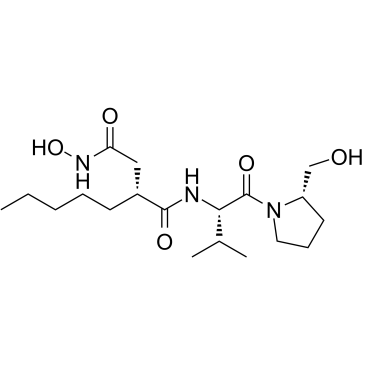
ACTINONIN structure
|
Common Name | ACTINONIN | ||
|---|---|---|---|---|
| CAS Number | 13434-13-4 | Molecular Weight | 385.498 | |
| Density | 1.1±0.1 g/cm3 | Boiling Point | N/A | |
| Molecular Formula | C19H35N3O5 | Melting Point | 137-139ºC | |
| MSDS | USA | Flash Point | N/A | |
Use of ACTINONINActinonin ((-)-Actinonin) is a naturally occurring antibacterial agent produced by Actinomyces. Actinonin inhibits aminopeptidase M, aminopeptidase N and leucine aminopeptidase. Actinonin is a potent reversible peptide deformylase (PDF) inhibitor with a Ki of 0.28 nM. Actinonin also inhibits MMP-1, MMP-3, MMP-8, MMP-9, and hmeprin α with Ki values of 300 nM, 1,700 nM, 190 nM, 330 nM, and 20 nM, respectively. Actinonin is an apoptosis inducer. Actinonin has antiproliferative and antitumor activities[1][2][3][4][5]. |
| Name | actinonin |
|---|---|
| Synonym | More Synonyms |
| Description | Actinonin ((-)-Actinonin) is a naturally occurring antibacterial agent produced by Actinomyces. Actinonin inhibits aminopeptidase M, aminopeptidase N and leucine aminopeptidase. Actinonin is a potent reversible peptide deformylase (PDF) inhibitor with a Ki of 0.28 nM. Actinonin also inhibits MMP-1, MMP-3, MMP-8, MMP-9, and hmeprin α with Ki values of 300 nM, 1,700 nM, 190 nM, 330 nM, and 20 nM, respectively. Actinonin is an apoptosis inducer. Actinonin has antiproliferative and antitumor activities[1][2][3][4][5]. |
|---|---|
| Related Catalog | |
| Target |
Ki: 0.28 nM (Peptide deformylase (PDF))[2], 300 nM (MMP-1), 1,700 nM (MMP-3), 190 nM (MMP-8), 330 nM (MMP-9)[3], and 20 nM (hmeprin α)[5] Apoptosis[1] Aminopeptidase M, Aminopeptidase N and Leucine aminopeptidase[1] |
| In Vitro | Actinonin inhibits cell growth in various human tumor cell lines. The IC50 of 4, 6.9, 12.8, 16.6, 27.4, 15.7 and 49.3 μM for Raji cells, MDA-MB-468 cells,PC3 cells, SK-LC-19 cells, Hela cells, HT-1080 cells and AL67 cells, respectively[1]. HsPDF is a critical target of actinonin and that the inhibition of this protein in the mitochondria leads to cell death in tumor cells. Actinonin treatment of cells led to a tumor-specific mitochondrial membrane depolarization and ATP depletion in a time- and dose-dependent manner[1]. Actinonin is a potent inhibitor of all three forms (Zn-, Ni-, and Fe-) of peptide deformylases from both S. aureus and E. coli bacteria. Under the assay conditions, the IC50 values for Actinonin are 90, 3, 0.8, and 11 nM for Zn-PDF (E. coli), Ni-PDF (E. coli), Fe-PDF (E. coli), and Ni-PDF (S. aureus), respectively[2]. Actinonin is active against Gram-positive bacteria, including S. aureus (MIC value of 8-16 µg/mL), Streptococcus pyogenes (MIC value of 8 µg/mL) and Streptococcus epidermidis (MIC value of 2-4 µg/mL). Actinonin is also active against fastidious Gramne-gative bacteria, such as H. influenzae (MIC value of 1-2 µg/mL), Moraxella catarrhalis (MIC value of 0.5 µg/mL), and Neisseria gonorrheae (MIC value of 1-4 µg/mL). Actinonin is very active against the H. influenzae acr (MIC value of 0.13 µg/mL) and E. coli acr (MIC value of 0.25 µg/mL) efflux pump mutants[2]. |
| In Vivo | Actinonin has been safely administered to mice as an antibiotic at doses up to 400 mg/kg. Actinonin does not appear to have significant toxicity to normal tissues, despite its antitumor activity in vitro. Remarkably, Actinonin exhibits significant antitumor activity when given i.p. or orally in a CWR22 human prostate tumor xenograft model in nude mice. During treatment, the animals show no signs of toxicity[1]. |
| References |
| Density | 1.1±0.1 g/cm3 |
|---|---|
| Melting Point | 137-139ºC |
| Molecular Formula | C19H35N3O5 |
| Molecular Weight | 385.498 |
| Exact Mass | 385.257660 |
| PSA | 118.97000 |
| LogP | 0.59 |
| Index of Refraction | 1.513 |
| Storage condition | −20°C |
CHEMICAL IDENTIFICATION
HEALTH HAZARD DATAACUTE TOXICITY DATA
|
| Personal Protective Equipment | Eyeshields;Gloves;type N95 (US);type P1 (EN143) respirator filter |
|---|---|
| Safety Phrases | S22-S24/25 |
| RIDADR | NONH for all modes of transport |
| WGK Germany | 3 |
| RTECS | RG9900700 |
|
Proteome-wide analysis of the amino terminal status of Escherichia coli proteins at the steady-state and upon deformylation inhibition.
Proteomics 15 , 2503-18, (2015) A proteome wide analysis was performed in Escherichia coli to identify the impact on protein N-termini of actinonin, an antibiotic specifically inhibiting peptide deformylase (PDF). A strategy and too... |
|
|
Quality control of mitochondrial protein synthesis is required for membrane integrity and cell fitness.
J. Cell Biol. 211 , 373-89, (2015) Mitochondrial ribosomes synthesize a subset of hydrophobic proteins required for assembly of the oxidative phosphorylation complexes. This process requires temporal and spatial coordination and regula... |
|
|
Concanavalin-A-induced autophagy biomarkers requires membrane type-1 matrix metalloproteinase intracellular signaling in glioblastoma cells.
Glycobiology 22(9) , 1245-55, (2012) Pre-clinical trials for cancer therapeutics support the anti-neoplastic properties of the lectin from Canavalia ensiformis (Concanavalin-A, ConA) in targeting apoptosis and autophagy in a variety of c... |
| Butanediamide, N-hydroxy-N-[(1S)-1-[[(2S)-2-(hydroxymethyl)-1-pyrrolidinyl]carbonyl]-2-methylpropyl]-2-pentyl-, (2R)- |
| Actinonine |
| MFCD00080257 |
| hylpropyl)carbamoyl) |
| (2R)-N-Hydroxy-N-{(2S)-1-[(2S)-2-(hydroxymethyl)pyrrolidin-1-yl]-3-methyl-1-oxobutan-2-yl}-2-pentylsuccinamide |
| (2R)-N-Hydroxy-N-{(2S)-1-[(2S)-2-(hydroxymethyl)-1-pyrrolidinyl]-3-methyl-1-oxo-2-butanyl}-2-pentylsuccinamide |