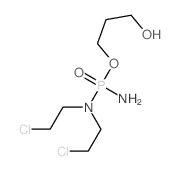
10159-53-2
| Name | phosphoramide mustard |
|---|---|
| Synonyms |
Phosphamide mustard
Friedman acid Phosphorodiamidic mustard Phosphoramide mustard amino-[bis(2-chloroethyl)amino]phosphinic acid Phosphorsaeure-amid-[bis-(2-chlor-aethyl)-amid] N,N-Bis(2-chloroethyl)phosphorodiamidic acid N,N-Bis-(2-chlor-aethyl)-diamidophosphorsaeure N,N-bis-(2-chloro-ethyl)-diamidophosphoric acid NLPD ASTA 5317 N-Lost-phosphorsaeurediamid [German] |
| Description | Phosphoramide mustard is the major metabolite for Cyclophosphamide (HY-17420), with anticancer activitiy. Phosphoramide mustard induces DNA adduct formation in ovarian granulosa cells, induces DNA damage and elicits the ovarian DNA repair response[1][2]. |
|---|---|
| Related Catalog | |
| Target |
DNA Alkylator[1] |
| In Vitro | Phosphoramide mustard causes cytotoxicity through forming cross-linked DNA adducts which inhibit DNA strand separation during replication[1]. Phosphoramide mustard destroys rapidly dividing cells by forming NOR-G-OH, NOR-G and G-NOR-G adducts with DNA, potentially leading to DNA damage[1]. Phosphoramide mustard (3-6 μM; 48 hours) reduces cell viability in rat spontaneously immortalized granulosa cells (SIGCs)[1]. Phosphoramide mustard (3-6 μM; 24-48 hours) induces DNA adduct formation[1]. Phosphoramide mustard (3-6 μM; 24-48 hours) induces ovarian DNA damage in rat ovaries[1]. Phosphoramide mustard increases DNA damage responses (DDR) gene (Atm, Parp1, Prkdc, Xrcc6, Brca1, Rad51) mRNA expression level[1]. Phosphoramide mustard (3-6 μM; 24-48 hours) increased DDR proteins[1]. Cell Viability Assay[1] Cell Line: SIGCs Concentration: 0.5 μM, 1 μM, 3 μM, 6 μM Incubation Time: 48 hours Result: Reduced cell viability at concentrations of 3 μM and higher. RT-PCR[1] Cell Line: SIGCs Concentration: 3 μM, 6 μM Incubation Time: 24 hours, 48 hours Result: Increased DDR gene mRNA expression levels. Western Blot Analysis[1] Cell Line: SIGCs Concentration: 3 μM, 6 μM Incubation Time: 24 hours, 48 hours Result: Generally increased DDR proteins. |
| In Vivo | Phosphoramide mustard (2.1-20.7 mg/kg; i.p.; daily; for 5 days) inhibits subcutaneous tumor growth in rats[2]. Phosphoramide mustard (59.4 mg/kg; i.v.) has a plasma disappearance half-life of 15.1 minutes[2]. Animal Model: Rat, subcutaneously implanted Walker 256 carcinosarcoma tumor[2] Dosage: 2.1 mg/kg, 4.8 mg/kg, 10.4 mg/kg, 20.7 mg/kg Administration: Intraperitoneal injection, once daily, for 5 consecutive days Result: Required to produce 50% inhibition of subcutaneous tumor growth with dose of 12 mg/kg. Animal Model: Rats[2] Dosage: 59.4 mg/kg (Pharmacokinetic Analysis) Administration: Intravenous injection Result: Has a disappearance half-life of 15.1 minutes in plasma. |
| References |
| Density | 1.474g/cm3 |
|---|---|
| Boiling Point | 363.5ºC at 760mmHg |
| Molecular Formula | C4H11Cl2N2O2P |
| Molecular Weight | 221.02200 |
| Flash Point | 173.6ºC |
| Exact Mass | 219.99400 |
| PSA | 76.37000 |
| LogP | 1.52540 |
| Vapour Pressure | 2.84E-06mmHg at 25°C |
| Index of Refraction | 1.525 |
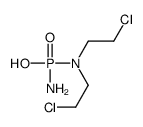
| Precursor 9 | |
|---|---|
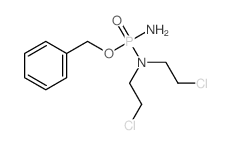
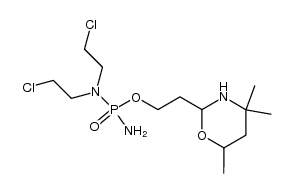
| DownStream 1 | |

![3-[amino-[bis(2-chloroethyl)amino]phosphoryl]oxy-1-phenyl-propan-1-one structure](https://image.chemsrc.com/caspic/352/100993-68-8.png)