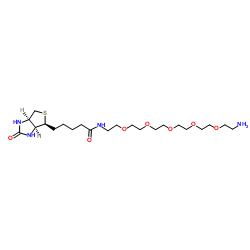
113072-75-6
| Name | N-(17-Amino-3,6,9,12,15-pentaoxaheptadec-1-yl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide |
|---|---|
| Synonyms |
1H-Thieno[3,4-d]imidazole-4-pentanamide, N-(17-amino-3,6,9,12,15-pentaoxaheptadec-1-yl)hexahydro-2-oxo-, (3aS,4S,6aR)-
N-(17-Amino-3,6,9,12,15-pentaoxaheptadec-1-yl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide |
| Description | Biotin-PEG5-amine is a biotin-labeled, PEG-based PROTAC linker that can be used in the synthesis of PROTACs[1]. |
|---|---|
| Related Catalog | |
| Target |
PEGs |
| In Vitro | PROTACs contain two different ligands connected by a linker; one is a ligand for an E3 ubiquitin ligase and the other is for the target protein. PROTACs exploit the intracellular ubiquitin-proteasome system to selectively degrade target proteins[1]. |
| References |
| Density | 1.2±0.1 g/cm3 |
|---|---|
| Boiling Point | 744.2±60.0 °C at 760 mmHg |
| Molecular Formula | C22H42N4O7S |
| Molecular Weight | 506.656 |
| Flash Point | 403.9±32.9 °C |
| Exact Mass | 506.277405 |
| LogP | -2.50 |
| Vapour Pressure | 0.0±2.5 mmHg at 25°C |
| Index of Refraction | 1.510 |